In this post I am sharing with you the Project Report that I have submitted as the work product for Google Summer of Code 2017. It summarizes all the blog posts written by me during this summer.
- Organisation - InterMine
- Github / @intermine
- Website / InterMine.org
- Project: Prototype a RESTful API Querying Neo4j Database
- Mentors:
- Sam / @sammyjava
- Daniela / @danielabutano
- Vivek / @vivekkrish
- Student: Yash Sharma / @yasharmaster
Table of contents
- Introduction
- Quick links
- How to use the code
- Metadata in Neo4j
- PathQuery to Cypher
- RESTful API
- Conclusion
Introduction
This document is for the purpose of review and for those interested to gain understanding about the work that I did for InterMine as a GSoC 2017 student developer. In this project we developed a prototype of InterMine which uses Neo4j database. The rest of the report describes the various tasks done during the summer.
Quick Links
How to use the code
Test the API via its Swagger UI
- Visit intermine-neo4jwebapp.herokuapp.com to test the API via its Swagger UI.
Host Server Locally
- Visit the project repo intermine/neo4j.
- Follow instructions given in README.md on how to host the server locally.
Metadata in Neo4j
Note - The metadata related code is in org.intermine.neo4j.metadata package.
In the present version of InterMine, the data model is stored in an external XML file. For each entity, its attributes, references and collections are stored in this XML file. For example, a part of the data model file which describes the BioEntity class looks like this.
<class name="BioEntity" is-interface="true">
<attribute name="primaryIdentifier" type="java.lang.String"/>
<attribute name="symbol" type="java.lang.String"/>
<reference name="organism" referenced-type="Organism"/>
<collection name="locatedFeatures" referenced-type="Location" reverse-reference="locatedOn"/>
<collection name="locations" referenced-type="Location" reverse-reference="feature" />
</class>While developing Neo4j prototype of InterMine, to reduce the dependency on external files, it was decided to store the schema in the Neo4j database itself. Since we can only store Nodes & Directed Relationships in Neo4j, therefore it brought up two issues:
- The data model must to be in the form of a graph itself so that it can be stored in the Neo4j database.
- The data model should represent all the existing Nodes in the database and the Relationships among them.
Since the data model is a graph and it stores information about the InterMine graph, we call it a metagraph. To create the schema/data-model for the database,
- Some sample data is loaded in Neo4j which adheres to the data model.
- As per the Nodes & Relationship of the sample data, metagraph is generated & stored in Neo4j.
- The data model can be retrieved and accessed using the Model class and its methods.
Metagraph Structure
Each node in the metagraph is assigned :Metagraph label. All the metagraph nodes are further classified into two types, NodeType and RelType. As the name suggests, each NodeType node represents a specific type of nodes and each RelType node represents a specific type of relationship in the IM graph. So each node has either :NodeType or :RelType label depending on which entity it represents.
NodeType
A metagraph node labelled :NodeType contains following two properties.
-
labels - A list containing the labels of the nodes that are represented by the
:NodeTypenode. For example, [“Gene”,”SequenceFeature”,”BioEntity”]. -
keys - A list containing the keys of all the properties exist amongst the nodes that are represented by the
:NodeTypenode. For example, [“primaryIdentifier”, “secondaryIdentifier”, “symbol”].
Each :NodeType node is uniquely identified by its labels property. So, for all the nodes in the IM graph which are labelled :Gene, :SequenceFeature, :BioEntity, there exists one :NodeType node in the metagraph which has its labels property set as [“Gene”,”SequenceFeature”,”BioEntity”].
RelType
A metagraph node labelled :RelType contains following two properties.
-
type - A string denoting the
typeof the relationships that are represented by the:RelTypenode. For example, “HOMOLOGUE_OF”. -
keys - A list containing the keys of all the properties exist amongst the relationships that are represented by the
:RelTypenode. For example, [“DataSet”].
Each :RelType node is uniquely identified by its type property. So, for all the relationships of type HOMOLOGUE_OF in the IM graph, there exists one :RelType node in the metagraph which has its type property set as “HOMOLOGUE_OF”.
Relationships in MetaGraph
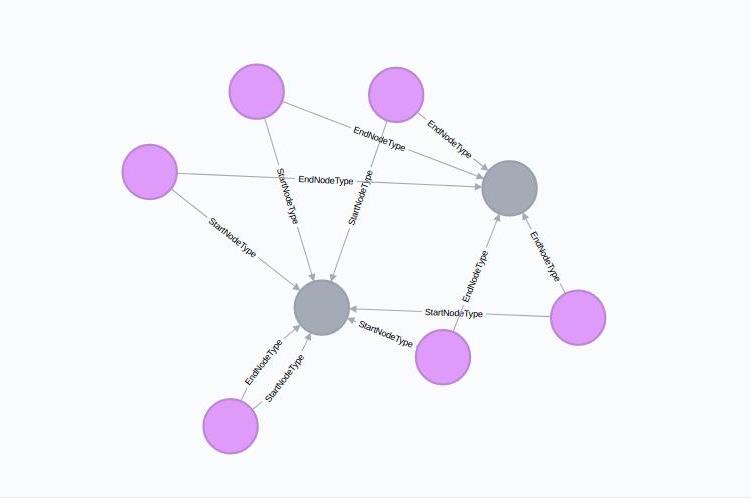
Metagraph should not only contain information about the properties of various entities in the IM graph but it should also store how different types of nodes are connected to each other. To represent this information, we make use of Neo4j relationships.
We know that each :RelType node represents a type of relationships that exist in the IM graph. Now, we create two outgoing relationships/edges from each :RelType node - :StartNodeType and :EndNodeType. These edges end on a :NodeType node.
Thus the metagraph path (a:RelType)-[:StartNodeType]->(b:NodeType) shows that the relationships represented by node a starts from the nodes represented by the node b. Same case follows for :EndNodeType.
PathQuery to Cypher
Note - The Path Query to Cypher conversion code is in org.intermine.neo4j.cypher package.
The InterMine graph is queried by the user via the API using Path Queries. On the other hand, Neo4j database is queried by Cypher Query Language. Therefore, we needed to convert the Path Query given by the user to Cypher Query. This conversion was the most difficult part of the project. Following is an example Path Query.
<query name="" model="genomic" view="Gene.primaryIdentifier" longDescription="" sortOrder="Gene.primaryIdentifier asc">
<constraint path="Gene.chromosomeLocation" op="DOES NOT OVERLAP">
<value>2L:14615435..14619002
</value>
</constraint>
</query>As you can see, various paths make up most of the Path Query. The information represented by paths is
- either returned as views,
- constrained to filter the data or
- used to order the results etc.
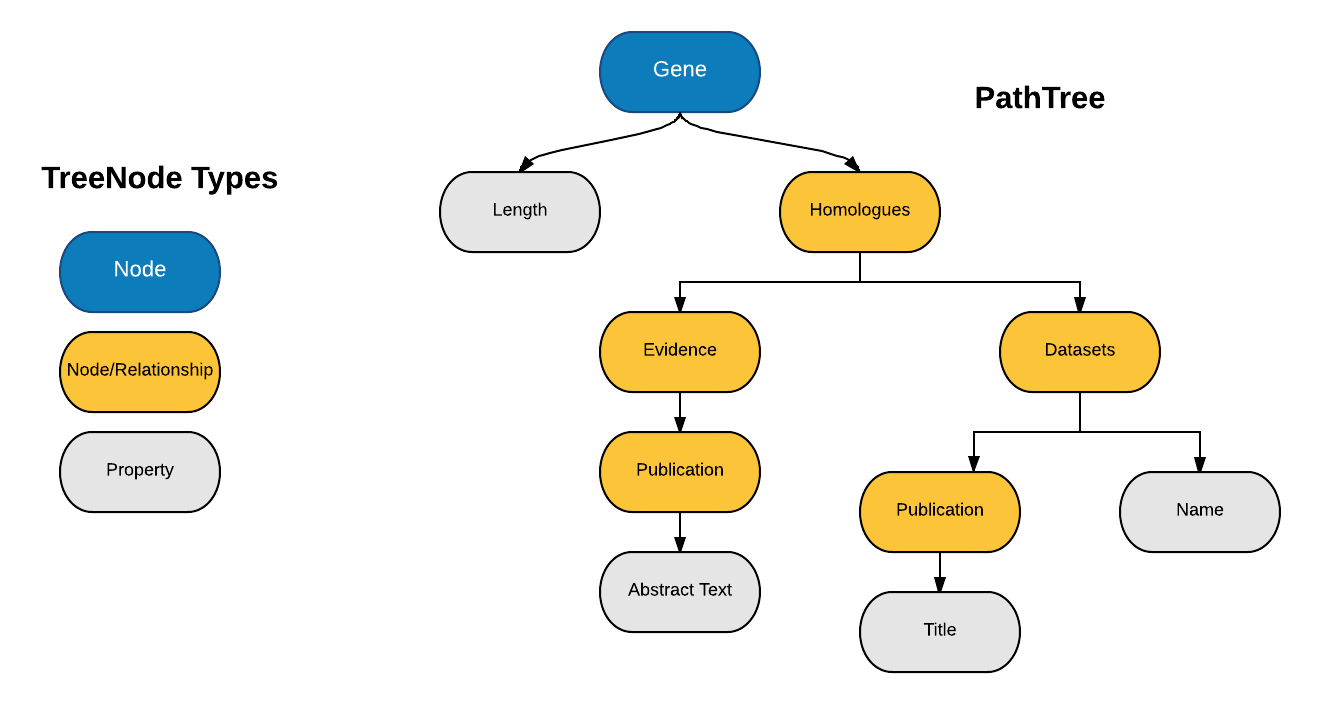
After pondering over the paths, I concluded that a Tree would be the best possible representation of all the paths of the Path Query. So, I created a data structure called a Path Tree and came up with an approach of Path Query to Cypher conversion which makes use of the Path Tree.
Roughly, the following approach was used by me to convert Path Query to Cypher.
1. Take a PathQuery object as input.
2. Retrieve all the Paths from the Views, Constraints & Sort Order.
3. Using all the Paths of PathQuery, create a PathTree such that
1. Each TreeNode represents a component of the path. For example, the path "Gene.pathways.identifier" forms three TreeNodes i.e. Gene, Pathways & Identifier.
2. Paths with common prefix have the same common ancestor.
3. Root TreeNode represents a Graph Node.
4. All Leaves of the PathTree always represent Graph Properties.
5. All other Internal TreeNodes can represent either Graph Nodes or Graph Relationships.
4. Generate & store a unique variable name to each Internal node of the PathTree.
1. This variable name will be used for referring that TreeNode in the cypher query.
2. For generating the variable name, we can separate each component of the path using underscores. For example, the variable name for "Gene.pathways.identifier" will be gene_pathways_identifier.
5. Use the PathTree & PathQuery to generate the cypher query
1. For creating the Match Clause, starting with the Root, recursively match each TreeNode of the PathTree,
1. If the TreeNode is Root, match the node itself. e.g. (gene).
2. If current TreeNode is a NODE,
1. If its parent is also a NODE, then fetch the Relationship Type from the XML data model file and create the match as (parentNode)-[relationshipFromXml]-(currentNode).
2. If the parent is a RELATIONSHIP, then fetch the grand parent from the PathTree and create match as (grandParentNode)-[parentNode]-(currentNode).
3. If current TreeNode is a RELATIONSHIP,
1. If current node does not have any children, then add match with an empty node as (parentNode)-[currentNode]-().
2. If current node has any children, then do nothing (they will be matched when recursion reaches the children).
2. For creating the WHERE clause,
1. For each constraint in the PathQuery, generate an equivalent Cypher constraint.
2. In the constraint logic of the PathQuery, replace the constraint code of each constraint with its equivalent Cypher constraint.
3. For creating the RETURN clause
1. For each view, get its path
2. Get the variableName of the last TreeNode of the path
3. Add variableNames separated by commas for each such variable
4. For creating ORDER BY clause,
1. For each Sort Order, get its path
2. Get the variableName of the last TreeNode of the path
3. Add variableName ASC/DESC separated by commas for each such variable
5. For handling JOIN operations in the PathQuery,
1. Add OPTIONAL MATCH clause in the query for corresponding paths.
7. Return the generated queryThe above approch is implemented in QueryGenerator class. After converting the Path Query given above, we will get the following Cypher query.
MATCH (gene :Gene),
(gene)-[gene_chromosomelocation:LOCATED_ON]-()
WHERE NOT (gene_chromosomelocation.start <= 14619002 AND gene_chromosomelocation.end >= 14615435 AND ()-[gene_chromosomelocation]-(:Chromosome {primaryIdentifier:'2L'}))
RETURN gene.primaryIdentifier
ORDER BY gene.primaryIdentifier ASCRESTful API
Note - The REST API code is in intermine-neo4jwebapp sub-project of the intermine/neo4j repository.
InterMine-Neo4j API
The InterMine-Neo4j API is developed using Jersey and is documented using Swagger. The InterMine-Neo4j API can be explored via Swagger UI at intermine-neo4jwebapp.herokuapp.com. There are two major endpoints in the API so far,
- query/cypher - This returns the Cypher equivalent of any Path Query.
- query/results - This endpoint returns the results of a Path Query by converting it into Cypher and running that Cypher against InterMine-Neo4j Graph.
API Response
We have kept InterMine-Neo4j API JSON response exactly similar to the InterMine API response. This would ensure that the applications that consume existing InterMine API can work seamlessly with the new API. The API response is being created manually in QueryResult class.
Conclusion
Contributing to InterMine this summer has been an amazing experience. It was my first time working on a project with a global team of experienced developers. I have got a chance to explore the latest technologies like Neo4j, Jersey, Gradle, Swagger etc.
I am grateful to my mentors Sam, Vivek and Daniela, for their guidance, direction and support throughout the summer. It is only because of their support and encouragement that I could complete this project. The entire InterMine community is welcoming and friendly. Overall it was a great learning experience!